Quantitating the Complete Human Proteome
Over 160,000 protein assays released through the Human SRMAtlas to allow the quantitation of essentially any Human protein 3 Bullets: ISB scientists collaborate with ETH Zurich to develop the Human SRMAtlas, a compendium of mass spectrometry assays for any human protein. ISB releases protein assay parameters freely to the scientific community for the ability to assay any human protein without restriction. Through the use of the ISB Human SRMAtlas, biomarker…
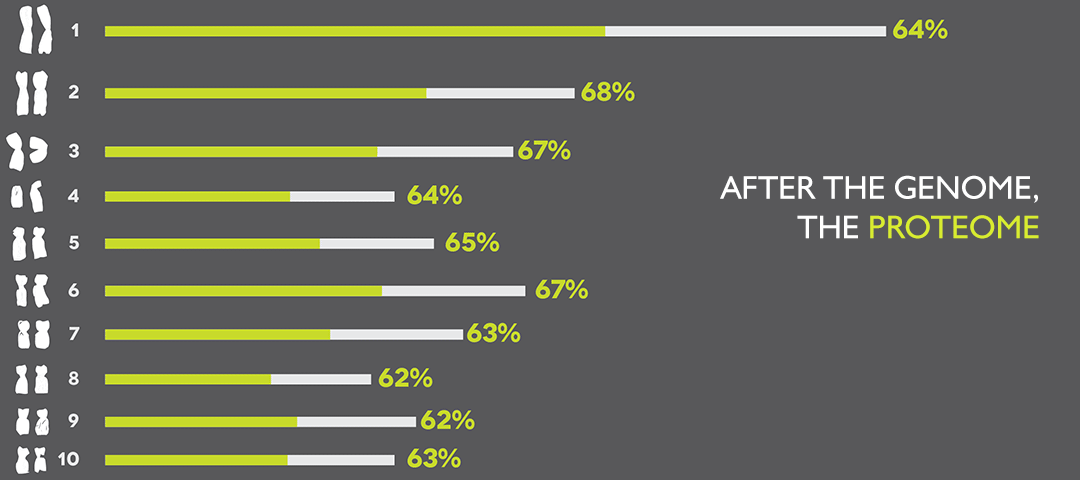
ISB’s PeptideAtlas Helps Advance the Study of Proteins
The Human Proteome Project expands on the work of the Human Genome Project. ISB is a leader in proteomics and developed PeptideAtlas, a project that has cataloged the proteins observed in thousands of proteomics experiments as an open resource for researchers everywhere. In a new report, ISB researchers show that PeptideAtlas now contains proteins from about 62 percent of human genes thought to encode proteins. By Terry Farrah Software Engineer,…

Let Us Tell You Everything We Know About Proteomics – Everything
3 Bullets: Proteomics experiments generate huge amounts of raw data, most of which cannot be easily shared or described in a publication. ISB researchers curate publicly accessible databases that allow researchers to share their data with the world and to use data others have collected. All data are analyzed in a consistent manner and results are presented via searchable, user-friendly web applications. By Dr. Kristian Swearingen Institute for Systems Biology…

ISB Releases Open-Source Software to Analyze Digital Fingerprint of Protein Data
3 Bullets: SWATH mass spectrometry, an emerging protein analysis technique being pioneered by ISB researchers, provides a digital fingerprint of all accessible proteins in a sample. The data generated by the SWATH technique are highly complex and require sophisticated computational tools in order to extract identities from a sea of data. ISB researchers have released a free, open source program that allows users to confidently identify and quantify proteins analyzed…



