-
Quantitating the Complete Human Proteome
Over 160,000 protein assays released through the Human SRMAtlas to allow the quantitation of essentially any Human protein 3 Bullets: ISB scientists collaborate with ETH Zurich to develop the Human SRMAtlas, a compendium of mass spectrometry assays for any human protein. ISB releases protein assay parameters freely to the scientific community for the ability to assay any human protein without restriction. Through the use of the ISB Human SRMAtlas, biomarker…
-

Systems Approach to Innovation Helps Extract More Data
Technology lag affects the study of proteins, which when quantified can indicate disease or wellness. ISB uses systems approach to innovate and transcend technology limitations. ISB co-develops new technique and software suite to increase protein detection rate. By Dr. Kristian Swearingen Scientific breakthroughs are often contingent on available technology. In the case of measuring proteins, the mass spectrometers that exist today aren’t fast enough to detect all the proteins that…
-
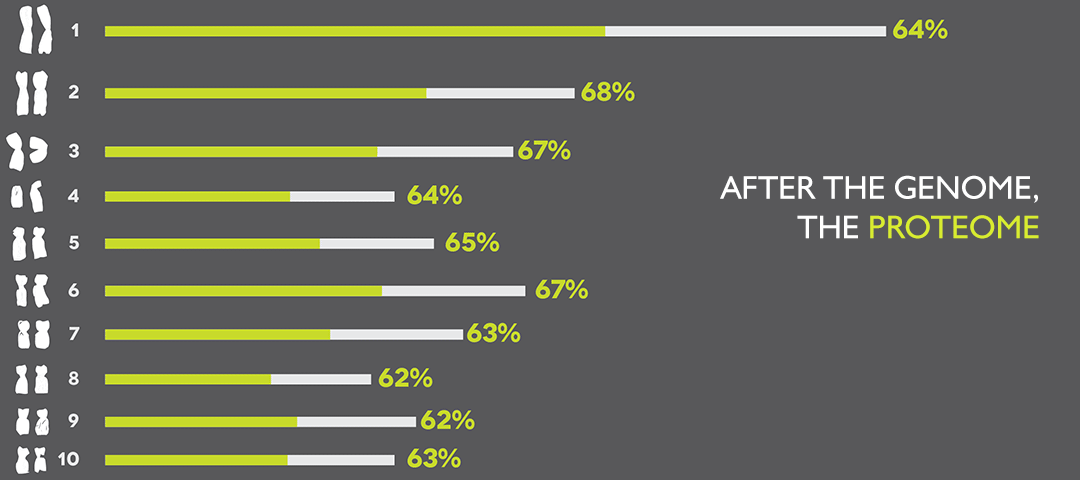
ISB’s PeptideAtlas Helps Advance the Study of Proteins
The Human Proteome Project expands on the work of the Human Genome Project. ISB is a leader in proteomics and developed PeptideAtlas, a project that has cataloged the proteins observed in thousands of proteomics experiments as an open resource for researchers everywhere. In a new report, ISB researchers show that PeptideAtlas now contains proteins from about 62 percent of human genes thought to encode proteins. By Terry Farrah Software Engineer,…
-
The Bug Stops Here: Arresting Malaria Parasites in the Liver Gives Immunity
ISB researchers collaborated with Seattle BioMed researchers to identify molecular building blocks required by malaria parasites to build cell membranes. Deleting key genes necessary for building cell membranes created a parasite that does not make the host sick and can’t be passed on through the blood. Treating mice with modified parasites gave them complete immunity against malaria. By Dr. Kristian Swearingen In a study published online in the journal Molecular…
-
New Protein Modification Critical to Growth of Tuberculosis Pathogen Found
Institute for Systems Biology and Seattle BioMed researchers collaborated and discovered a new protein post-translational modification in the human pathogen Mycobacterium tuberculosis. Post-translational modifications are essential mechanisms used by cells to diversify protein functions and ISB scientists identified the rare phosphorylated tyrosine post translational modification on Mycobacterium tuberculosis proteins using mass spectrometry. Inhibiting phosphotyrosine modified amino acids in Mycobacterium tuberculosis severely limits the growth of this widespread deadly pathogen. By…
-

A New Approach to Identifying How the Deadly Dengue Virus Multiplies
3 Bullets Dengue virus is the most prevalent mosquito-borne virus worldwide, infecting an estimated 400 million people per year and causing about 25,000 deaths. It’s necessary to understand the molecular mechanisms of dengue replication in order to develop an effective treatment. Researchers at ISB and Seattle BioMed developed a novel approach for identifying host proteins that associate with dengue replication machinery. By Thurston Herricks Dengue virus (DENV) infects approximately 400…
-

Let Us Tell You Everything We Know About Proteomics – Everything
3 Bullets: Proteomics experiments generate huge amounts of raw data, most of which cannot be easily shared or described in a publication. ISB researchers curate publicly accessible databases that allow researchers to share their data with the world and to use data others have collected. All data are analyzed in a consistent manner and results are presented via searchable, user-friendly web applications. By Dr. Kristian Swearingen Institute for Systems Biology…
-

Identifying markers of healthy skin development
3 Bullets The barrier function of skin is integral to personal well-being and is associated with several widespread diseases such as eczema and psoriasis. ISB and Procter & Gamble researchers used human skin grown in the lab to measure changes in protein levels as the skin matures. The results of this study provide many new markers for healthy skin development. By Dr. Kristian Swearingen and Dr. Jason Winget In a…
-

ISB Releases Open-Source Software to Analyze Digital Fingerprint of Protein Data
3 Bullets: SWATH mass spectrometry, an emerging protein analysis technique being pioneered by ISB researchers, provides a digital fingerprint of all accessible proteins in a sample. The data generated by the SWATH technique are highly complex and require sophisticated computational tools in order to extract identities from a sea of data. ISB researchers have released a free, open source program that allows users to confidently identify and quantify proteins analyzed…
-
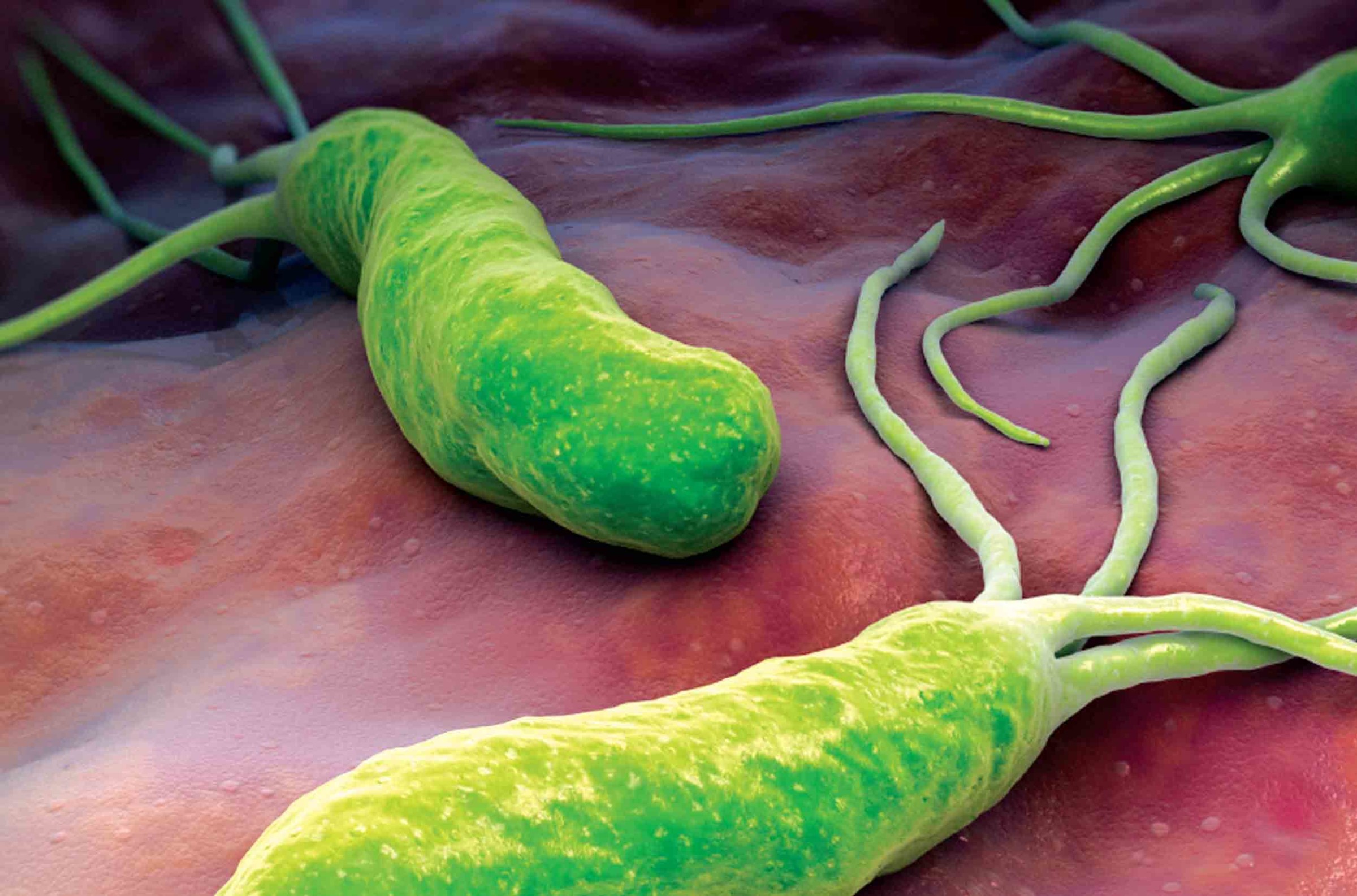
Ötzi the Iceman: Helicobacter pylori Pathogen Found in Stomach Contents
Institute for Systems Biology collaborates with researchers worldwide to study pathogens in the stomach content and microbiome of the 5300 year old European Copper Age glacier mummy “Ötzi” and discovers a Helicobacter pylori pathogen genome. Ötzi harbored a nearly pure Asian-origin bacterial population of H. pylori providing key information as to population migration into Europe over the last few thousand years. Supported by proteome information gathered at ISB, the Iceman’s…
-
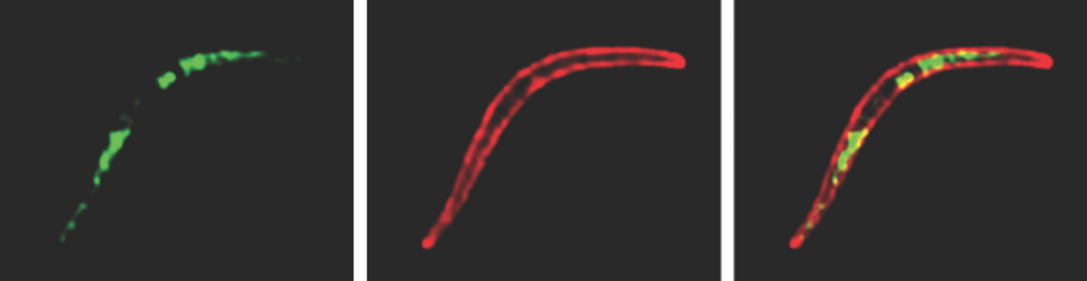
Scratching the Surface of the Malaria Parasite
Surface coat proteins on Plasmodium, the parasite that causes malaria, provide new targets for a malaria vaccine. 3 Bullets: ISB scientists collaborate with scientists from Center for Infectious Disease Research and Johns Hopkins University to identify new proteins on the surface of the malaria parasite Plasmodium. The research aims to provide new targets for a malaria vaccine. It is discovered that some Plasmodium surface proteins have sugar attachments that can…
-

FAIMS
FAIMS–Kristian Swearingen, Michael Hoopmann, and Robert Moritz (PI) What is FAIMS? High Field Asymmetric Waveform Ion Mobility Spectrometry (FAIMS) is an established technology with a myriad of potential applications for augmentation of mass spectrometry. Although discovered and characterized for use with mass spectrometry by Gorshkov in the early 1980’s, FAIMS technology has recently seen a marked increase in interest and innovation through the work of Roger Guevremont, Alexandre Shvartsburg, Richard…
Privacy Overview
This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.



